Table of Contents
Disaggregation of crop Areas
Files:
%curdir%/capdis/disyield.gms %curdir%/capdis/disyield_sets.gms %curdir%/capdis/m_hpdCropSpat.gms %datdir%/capdishsu/pesetagrid_fractionfsu.gdx %datdir%/capdishsu/irriShare2000fsu.gdx
The principle of the distribution of crop areas is based on few constraints only: full exhaustion of available ares for each spatial unit, vertical consistency, and primacy of land stability.
Vertical consistency means that the sum of each land use type over all spatial units recovers available land use at the higher spatial level. As available information are not (necessarily) geo-referenced, the allocation of a given statistical information to a spatial unit is associated with uncertainty. For example, a farmer with the residence in region A can have land also in regions B and C, but will declare them together, and they will be allocated to her residence (region A). Accordingly, there is some blurring in particular at boundaries and this is accounted for in the methodology: also at the high disaggregation level, the land uses are principally to be interpreted as ‘Land owned by a farmer with residence in this spatial unit’.
Primacy of land stability means that if there is no indication (i. e. new observation, policy restricting previous land distributions, …) it is more likely that the spatial pattern remains similar to the previous (prior) pattern. Therefore, once a likely distribution of land and livestock has been determined on the basis of high-resolution FSS statistics, the model tries to stay as close as possible to this distribution. This is achieved with penalty factors that are activated as soon as the estimated land use area deviated from the prior values, assigning a higher penalty for deviations of permanent crops and forests, and very high penalties of a land use is estimated in a spatial unit where it didn’t exist in the prior’s data base.
The disaggregation model m_hpdCropSpat is described in Section Simulation model m_hpdCropSpat.Section Data sets describes the required input data, and Section Data preparation describes the preparation of the input data for their use in hpdCropSpat.
Simulation model m_hpdCropSpat
Equation 1 RULEVL_
The levels assigend to the FSUs must recover the given NUTS II area
both NUTS2 level and FSU level in 1000 ha
rur = grid cell in mode LAPM, rur = NUTS2 in all other modes
RULEVL_(rur,curact) ..
SUM(regmap(rur,cur%spatunit%),v_levlCon(rur,cur%spatunit%,curact))
=E= p_nutsLevl(rur,curact);
\begin{equation} A_{c^*,r}=\sum_h\{a_{c^*,h}\} \end{equation}
\(a_{c^*,h}\) = Area [parameter, km2] cultivated with crop c or covered by ‘other land’ use excluding forest in spatial unit h
\(A_{c^*,h}\) = Area [parameter, km2] cultivated with crop c or covered by ‘other land’ use excluding forest in region r
Equation 2 ADDUPGRID_
Due to several reasons it could be impossible to distribute all agricultural area under the given constraints of total available area in the spatial units (net of the area of ‘nogo’ units and forest area). In order to enable a feasible solution, an error term is introduced that allows the units to slightly shrink or grow. The reason:
- Statistical data are not (necessarily) geo-referenced; i.e. an area of crops (or a livestock) might be assigned to one unit/grid cell because this is where the farm is registered rather than the physical location of crop/livestock
- Uncertainties in the data
- Inconsistencies of data sources (i.e. FSS agricultural statistics, Corine Land Cover data, CAPRI regional statistics)
- – The FSU area must be exhausted, but the variable v_%spatunit%SizeChg
allows some flexibility if needed.
ADDUPGRID_(rur,cur%spatunit%) $ p_levlunit(rur,cur%spatunit%,"area") .. SUM(curact,v_levlCon(rur,cur%spatunit%,curact)) =E= v_%spatunit%SizeChg(rur,cur%spatunit%)*p_levlunit(rur,cur%spatunit%,"area");
\begin{equation} a_h⋅\epsilon_{a,h}=\sum_{c^*}\{a_{c^*,h}\} \end{equation}
\(a_h\) = Area [parameter, km2] of spatial unit h
\(a_{c^*,h}\) =Area [parameter, km2] cultivated with crop c or covered by ‘other land’ use excluding forest in spatial unit h
\(\epsilon_{a,h}\) = Error term, allowing a spatial unit to shrink or grow slightly in order to enable a feasible disaggregation of statistical data.
Equation 3 PDF_
The most likely solution is obtained with the ‘Highest Posterior Density’ method. A penalty function calculates deviations from prior information, applying uncertainties
- A random re-allocation of crops should be avoided. Therefore, a penalty is given with increasing deviation from the prior distribution to ensure stability in the time series
- In particular the ‘appearance’ of crops in spatial units where they have not been observed in the prior data should be restricted (disagg(“penelizenewcrops”))
- The error term for the area of the spatial units should be kept at a minimum (disagg(“penalizesizechange”))
PDF_ ..
# Scale density value a good couple of magnitudes for numerical reasons.
v_hpd
*[SUM((curact,regmap(rur,cur%spatunit%)) $ p_levlStde(cur%spatunit%,curact),
p_levlunit(rur,cur%spatunit%,"area"))
+SUM(regmap(rur,cur%spatunit%) $(v_%spatunit%SizeChg.LO(rur,cur%spatunit%) NE
v_%spatunit%SizeChg.UP(rur,cur%spatunit%)),
p_levlunit(rur,cur%spatunit%,"area"))
]
=E=
# hsu area-weighted mean square of the deviation from prior mean area, scaled by its stdev
(SUM((regmap(rur,cur%spatunit%),curact),p_levlunit(rur,cur%spatunit%,"area")
* SQR( (v_levlCon(rur,cur%spatunit%,curact)-p_levlunit(rur,cur%spatunit%,curact))
*( 1 $ p_levlunit(rur,cur%spatunit%,curact)
+ disagg("penalizenewcrops") $ (not p_levlunit(rur,cur%spatunit%,curact)))
/max(1e-3,
$$ifi %MODE%==LAPM p_levlStde(cur%spatunit%,curact)
$$ifi NOT %MODE%==LAPM 1
)
) \\
)/SUM((regmap(rur,cur%spatunit%),curact) \\
$p_levlStde(cur%spatunit%,curact),p_levlunit(rur,cur%spatunit%,"area")) \\
)$sum((regmap(rur,cur%spatunit%),curact),p_levlStde(cur%spatunit%,curact)) \\
# penalty for deviation from hsu area \\
+(SUM(regmap(rur,cur%spatunit%), \\
disagg("penalizesizechange")*p_levlunit(rur,cur%spatunit%,"area")*SQR((v_%spatunit%SizeChg(rur,cur%spatunit%)-1))) \\
/SUM(regmap(rur,cur%spatunit%), p_levlunit(rur,cur%spatunit%,"area")) \\
)$SUM(regmap(rur,cur%spatunit%),p_levlunit(rur,cur%spatunit%,"area")) \\
; \\
Model parameters
Some model parameters can be set by the user through the CAPRI GUI.
They are collected in the parameter ‘disagg’.
set disaggcontrol / mincropshare "Minimum allowed cropshare per HSU" relcropshare "Defines heterogeneity of crop shares for a crop per HSU" relstdefix "Relative standard deviation, predefined if land needs 'to be fixed'" relstdeperm "Relative standard deviation, predefined for permanent crops" relstdeothe "Relative standard deviation, predefined for other land (large to avoid that other land pushes agriland around)" penalizenewcrops "multiply deviations for crops predicted where they haven't been before" penalizesizechange "multiply deviations for total HSU unit size" * --- scalars controlling livestock disaggregation weightRUMIfodduaar "Weighting between fodds and uaar to distribute initial RUMI numbers" weightMONOcereuaar "Weighting between cereals and uaar to distribute initial NRUMI numbers" minLSUdens "Minimum density for LSU allowed to not have them everywhere... " # managing crop residues minmcactSurs #Miniumum surplus as compared to average over all crops maxmcactSurs #Maxiumum surplus as compared to average over all crops rangemcactSurs #Range of sursoi (max/average) below which the high sursoi are not reduced /;
- Stability of forests – disagg(“relstdefix”)
Forests cannot easily be ‘displaced’ and are likely to remain rooted as given in the land cover data sets. So far, estimations of changes of forest areas at the regional level are not included in the disaggregation procedure.
The default value used for disagg(“relstdefix”) = 0.01
The lower the value the higher becomes the penalty if the estimates are deviating from the priors.
- Stability of permanent crops disagg(“relstdeperm”)
Permanent crops are long-term investments and require time to grow. Displacement of permanent crops is slow.
The default value used for disagg(“relstdeperm”) = 0.05.
- Coefficient of variation for ‘other land uses’ disagg(“relstdeothe”)
‘Other’ area is a lump of all non-agricultural areas. We consider this area as relatively flexible.
The default value used for disagg(“relstdeothe”) = 1.
- Penalization for new crops in spatial units disagg(“penalizenewcrops”)
New crops ‘appearing’ in spatial units, if they have not been in the priors data set, are penalized. The penalization factor is a multiplicator of the squared deviation from the prior. Thus, the higher the factor the higher becomes the penalty.
The default values used for disagg(“penalizenewcrops ”)=2.0;
- Penalization of area changes of spatial units disagg(“penalizesizechange”)
The default values used for disagg(“penalizesizechange”)=2.0;
- Minimum crop share allowed in the spatial unit disagg(“mincropshare”)
The minimum crop share which is allowed in the spatial unit χ_(min ) is used to calculate the lowest allowed crop share, in combination with the minimum relative crop share defining the level of spatial heterogeneity for a crop. See section 7.4.3.5 
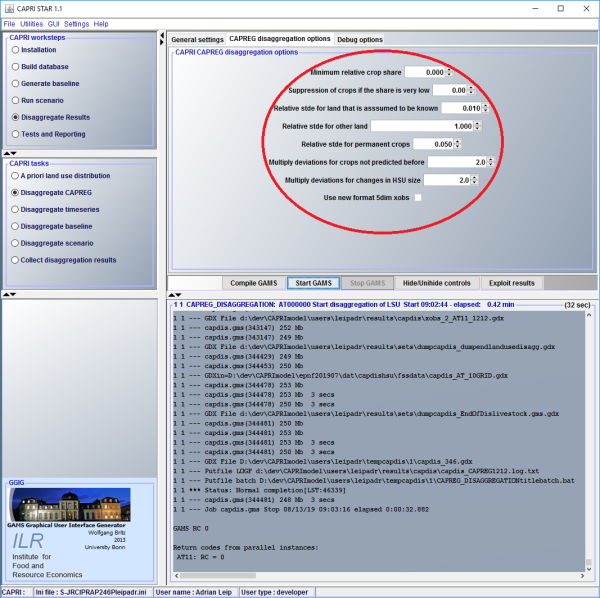
\(χ_{min}\) can be set through the CAPRI GUI (tab CAPREG disaggregation options – “Suppression of crops if the share is very low”)
By default, \(χ_{min}\)is set to zero.
- Minimum relative crop share disagg(“relcropshare”)
The minimum relative crop share defining the level of spatial heterogeneity for a \(χ_{rel}\) is used to calculate the lowest allowed crop share, in combination with the m crop minimum crop share which is allowed in the spatial unit (see section 7.4.3.5 
\(χ_{rel}\) can be set through the CAPRI GUI (tab CAPREG disaggregation options – “Minimum relative crop share”)
By default, \(χ_{rel}\) is set to zero.
Defining bounds for the land use distribution model
Bounds for size changes of the total area of the spatial units
v_%spatunit%SizeChg.L (rur,cur%spatunit%) $p_temp3dim(rur,cur%spatunit%,"crops")= 1; v_%spatunit%SizeChg.UP(rur,cur%spatunit%) $p_temp3dim(rur,cur%spatunit%,"crops")= 1.1; v_%spatunit%SizeChg.LO(rur,cur%spatunit%) $p_temp3dim(rur,cur%spatunit%,"crops")= 0.9; v_%spatunit%SizeChg.UP(rur,cur%spatunit%) $(p_nutslevl(rur,"AREAcorr") and p_temp3dim(rur,cur%spatunit%,"crops"))= 2.0; v_%spatunit%SizeChg.LO(rur,cur%spatunit%) $(p_nutslevl(rur,"AREAcorr") and p_temp3dim(rur,cur%spatunit%,"crops"))= 0.5; $$ifi %MODE%=="LAPM" v_%spatunit%SizeChg.UP(rur,cur%spatunit%) $p_temp3dim(rur,cur%spatunit%,"crops")= max(1.1,p_nutslevl(rur,"AREAcorr")); $$ifi %MODE%=="LAPM" v_%spatunit%SizeChg.LO(rur,cur%spatunit%) $p_temp3dim(rur,cur%spatunit%,"crops")= 1/max(1.1,p_nutslevl(rur,"AREAcorr"));
To enable the solver to find feasible solutions even in difficult situations, it is possible to expand or shrink the total area of the spatial units. This is in consistence with the definition of the data compiled by the statistical offices which link the area to residence to the farmer rather than to the geographic location of each field.
Generally, we limit this area-change to plus/minus 10% of the original size.
In cases where inconsistencies between data sets have already been identified (see 0) a higher degree of flexibility is allowed (factor 2) as it is not known in which spatial unit the inconsistency originated.
Only in the task ‘A priori land use distribution’ the degree of flexibility is calculated as a function of the correction that had to be applied to the regional area.
The bounds for the area-size change are hard-coded and can not be changed by the user.
Setting standard deviations
Data do not come with any level of uncertainty attached, and there is no a priori information on what spatial distribution is more likely than any other.
Therefore, the uncertainty in the estimates is ‘guessed’ based on crop groups.
Other options tested (all standard deviations equal or scaling prior estimates to a plausible range) are currently not used. The standard deviations are only set at the first task. In subsequent tasks, the standard deviations of the priors are used and ‘gap-filled’ if necessary.
$set changelapmstdev bygroups
$iftheni.std %changelapmstdev%=="scalestdevs"
$elseifi.std %changelapmstdev%=="allone"
$elseifi.std %changelapmstdev%=="bygroups"
p_levlstde(cur%spatunit%, %croptp%)
* $ p_levlstde(cur%spatunit%, %croptp%)
= 0.5;
p_levlstde(cur%spatunit%, %croptp%)
* $ p_levlstde(cur%spatunit%, %croptp%)
= 0.001 $sum(fssact2groups(%croptp%, "FORE"), 1)
+ 0.50 $sum(fssact2groups(%croptp%, "CERE"), 1) # Cereals incl those likely in rotation: Assumptions market oriented might change relatively quickly if price is correct
+ 0.25 $sum(fssact2groups(%croptp%, "FODD"), 1) # Fodder crops: roof, ofar, lgras. Assumption: link to livestock which do not shift around so quickly
+ 0.25 $sum(fssact2groups(%croptp%, "OILS"), 1) # Oil crops: rape, sunflower, soya. Assumption: relatively sticky
+ 0.15 $sum(fssact2groups(%croptp%, "VEGE"), 1) # Vegetables: flower, pulses, potatoes, sugar beet, ... but also tobacco, text etc. Assumption: require often infrastructure (greenhouse) so longer investments required
+ 0.05 $sum(fssact2groups(%croptp%, "TREE"), 1) # Permanent crops: olives, nurseries, fruit and nuts trees, vinyards. There are permanent
+ 0.05 $sum(fssact2groups(%croptp%, "PERM"), 1) # Permanent crops: olives, nurseries, fruit and nuts trees, vinyards. There are permanent
+ 0.80 $sum(fssact2groups(%croptp%, "REST"), 1) # Assumptions: can be easily pushed around
;
Data sets
Update pending
FSS 2010 data at nested grid levels
FSS 2010 data, gap-filled at 10km-NUTS3 overlay
Forest map
Data preparation
Re-mapping from FSS crops to CAPRI crops
# Convert to CAPRI activities (posteact) #As grids and NUTS are not always consistent - use the grid-HSUs to fill with crops p_hsufraction(curgrid,%spatunit%_all)=p_hsu_grid10n23(%spatunit%_all,curgrid,"fracHSU"); grid10n23_hsu(curgrid,%spatunit%_all)$p_hsufraction(curgrid,%spatunit%_all)=yes; cur%spatunit%(%spatunit%_all)=yes$sum(grid10n23_hsu(curgrid,%spatunit%_all),1);
Intersecting FSS 2010 10km x NUTS3 to FSU
The land use distribution model works at the spatial intersection of the prior and posterior spatial units. Historically (when CAPDIS was based on HSU) this intersection was an area determined by the fraction of the HSU that lies within different FSS-admin grid cells. This fraction \(f_{hg}\in[0,1]\).
However after the update to the FSU, each spatial unit was fully in one FSS-admin grid cell only, thus \(f_{hg}\in\{0,1\}\). The following text is therefore relevant only if CAPDIS is run with ‘old’ HSU.
# Work on the intersection between grid and HSU (Line 585 ff) p_levlunit(%region%,cur%spatunit%,"AREA") =p_hsufraction(%region%,cur%spatunit%)*p_hsu(cur%spatunit%,"area"); " Work on intersection of HSU and grid for crops"' '" "' # Distribute crops over intersected units –and scale to total area (should be already but not always is...) p_levlunit(%region%,cur%spatunit%,%croptp%) =p_hsufraction(%region%,cur%spatunit%)*p_levlmean(cur%spatunit%,%croptp%); # Scale all areas such that the sum becomes the AREA of the unit (in case the HSU had to be split) # - if LAPM predictions are consistent with total area p_temp3dim(%region%,cur%spatunit%,"allarea") =sum(%croptp%$(not sameas(%croptp%,"FORE")),p_levlunit(%region%,cur%spatunit%,%croptp%)); p_levlunit(%region%,cur%spatunit%,%croptp%) $(p_temp3dim(%region%,cur%spatunit%,"allarea") and not sameas(%croptp%,"FORE")) =p_levlunit(%region%,cur%spatunit%,%croptp%)*(p_levlunit(%region%,cur%spatunit%,"AREA")- p_levlunit(%region%,cur%spatunit%,"FORE")) /p_temp3dim(%region%,cur%spatunit%,"allarea");
In order to ensure consistency of total area between the prior and the posterior data sets, all data are re-mapped into their intersection. This is achieved with the fraction of the spatial units of one layer that is part of a unit of the second spatial layer:
\begin{equation} a_{hg}=\sum_{h,g}\{a_h\cdot f_{hg} \} \end{equation}
\(a_{hg}\) = Area [parameter, km2] of unit u intersecting spatial unit h and grid cell g.
\(a_h\) = Area [parameter, km2] of spatial unit h
\(f_{hg}\) = Fraction of spatial unit h, which is covered by grid cell g
Cultivated crop areas are re-mapped to the intersecting units proportionally to the area fraction, assuming homogeneous distribution of each crop within each spatial unit h.
The LAPM predictions are not constrained to exhaust the total available area. However, this is a required characteristic in CAPRI. Therefore, the land use areas (crops, forest land and ‘other’ area) are scaled so that their sum matches the total available area. As forest areas are obtained from a data set, which is assumed to be of high precision, forest areas are excluded from scaling.
\begin{equation} a_{c^o,hg}=\sum_{h,g} \left\{ a_{c,h} \cdot f_{hg} \cdot \frac{a_{hg} - a_{forest,hg}}{\sum_{c^{o^\prime}} a_{c^{o^\prime}}} \right\}, \end{equation}
\(c^*\) = Land use. \(c^*\in\{c,other \; land\}\)
\(a_{c^o,hg}\) = Area [parameter, km2] cultivated with crop c or covered by ‘other land’ use excluding forest in unit u intersecting spatial unit h and grid cell g.
\(a_{c^o,h}\) = Area [parameter, km2] cultivated with crop c or covered by ‘other land’ use excluding forest in spatial unit h
\(f_{hg}\) = Fraction of spatial unit h, which is covered by grid cell g
Note that this re-mapping is done in each step, however it affects only the step ‘A priori land use distribution’ which is constrained by data for the intersections of the FSS-10km grid cells with NUTS3 regions. These units are not aligned with the spatial units (HSU) for two reasons:
- HSU are aligned with a regular grid of 0.25° x 0.25° but not to a grid of 10 km x 10 km
- Even though HSU are aligned with a NUTS3 administrative region layer, changes in the definition of NUTS3 regions over time create shifts in the boundaries
In all other steps, the constraining data set is taken from CAPRI NUTS2 regions to which all spatial units are nested to and \(f_{hr}=1\forall h,r\).
The same holds if disaggregation is done into the FSU units, which are part of exactly one FSS grid cell.
Dealing with FSS grid cells with too much crops
Line 613ff p_temp3dim(%region%,"AREA","<0")$(p_nutslevl(%region%,"AREA") and (p_nutslevl(%region%,"OTHER")<0)) =(p_nutslevl(%region%,"CROPS")+p_nutslevl(%region%,"FORE"))/p_nutslevl(%region%,"AREA"); p_nutslevl(%region%,"AREAcorr")=p_temp3dim(%region%,"AREA","<0"); # Rescale total and crop area of units if the total area in the %region% had to be changed. p_levlunit(%region%,cur%spatunit%,"AREA")$(p_temp3dim(%region%,"AREA","<0")) =p_levlunit(%region%,cur%spatunit%,"AREA")*p_temp3dim(%region%,"AREA","<0"); p_levlunit(%region%,cur%spatunit%,"OTHER")$(not p_temp3dim(%region%,"AREA","<0")) =max(0,p_levlunit(%region%,cur%spatunit%,"AREA")- sum(%croptp%,p_levlunit(%region%,cur%spatunit%,%croptp%)));
Farm structure surveys collect data on crop areas and allocate them to the geographic location where the farmer resides. Therefore, it is not excluded that a spatial unit (grid cell) has more crop area than the cell is large. It is not possible to ‘correct’ those allocations.
CAPRI works with an area-consistent approach, thus the total area available must be exactly matching the sum of the areas used for different purposes. As a re-allocation of ‘surplus’ crop areas is not possible, we inflate the area of spatial units h so that all forest and crop areas can be accommodated. The area of ‘other’ land uses is adapted to ensure coherence.
\begin{equation} a_{hg} \leftarrow a_{hg} \cdot \frac{a_{g,forest} + \sum_c a_{g,c}}{a_g} \end{equation}
\begin{equation} a_{hg, other} = a_{hg} - a_{hg,forest} - \sum_c a_{hg,c} \end{equation}
Note that this ‘manipulation’ of the data is needed to avoid any potential infeasibilities in the land use disaggregation model maintaining the relevant information from the different data sets:
- The total crop areas data collected in the FSS
- The heterogeneity of crop areas (‘suitability’) as modelled with the LAPModel. This concerns both the spatial heterogeneity within a region across spatial units, as well as the relative abundance of different crops in a single spatial unit.
Adding previously unobserved crops
It might well be that crops occur in a grid cell or region which were not predicted or which had not been observed ‘before’ (e.g. when moving from ex-post to ex-ante simulation). In this case prior estimates of the distribution need to be developed.
This is done on the basis of ‘similarity’ assuming that similar crops have similar preferences for natural conditions (or available infrastructure) and a similar spatial heterogeneity.
This ‘gap-filling’ is done in three hierarchical steps:
1. Average crop area of similar crops in the same spatial unit as defined in the set mactgroups:
set mactgroups(sgroups,*) "Groups with similar crops - LAPM activities" / CERE.(BARL,SWHE,DWHE,LMAIZ,OATS,RYEM,OCER,PARI) VEGE.(TOMA,OVEG,SUGB,POTA,PULS,FLOW) FODD.(ROOF,OFAR,LGRAS) OILS.(SOYA,LRAPE,SUNF) FORE.(FORE) TREE.(APPL,CITR,OFRU,NURS,NUTS,LOLIV,LVINY)/;
2. If there are no ‘similar’ crops in the region or grid cell, the average area of all available crops is used.
3. If there is still no prediction of the in the spatial unit, the same crop area is given to the prior estimates in all spatial units in the region/grid cell.
Checking availability of standard deviations
May become obsolete for the CAPDIS modules of the CAPRI stable release versions following STAR 2.4
The LAPModel provides not only prediction for crop shares for each HSU, but at the same time also an estimate for the standard deviation of each estimate. These standard deviations are ‘carried’ on throughout the CAPRI disaggregation steps.
The procedures described above however made evident that some crops might appear in spatial units where they have not been observed before; obviously, an estimate of the standard deviation must be provided as well.
If the crops have been observed in other spatial units of the region or grid, the maximum relative standard deviation is used. If the crop has not been observed in the whole region or grid, the maximum standard deviation over all observed crops is used. Still missing standard deviations are assumed to be 100%.
Standard deviation for ‘other land uses’ are also set to a default of 100%, but can be modified by the user (through the CAPRI GUI).
Preparation for specific applications
Lines 690ff
p_temp3dim(%region%,"loshare",%croptp%)$ p_nutslevl(%region%,%croptp%)
= max(disagg("mincropshare"),p_temp3dim(%region%,"share",%croptp%)*disagg("relcropshare"));
p_levlunit(%region%,cur%spatunit%,%croptp%)
$((p_levlunit(%region%,cur%spatunit%,%croptp%)/sum(%croptp%1,p_levlunit(%region%,cur%spatunit%,%croptp%1))
<p_temp3dim(%region%,"loshare",%croptp%)) and not (sameas(%croptp%,"other")))
=0;
For some applications, the focus is on the analysis of dominant crops in each spatial unit, for example, when linking the result of the disaggregation with process-based crop models. This is done on the basis of two parameters:
\begin{equation} a_{hc}= 0 \leftarrow \frac {a_{h,c}} {\sum_{c^{\prime}}a_{h,c^\prime}} \lt max \left[ χ_{min},\frac{a_{r,c}}{\sum_{c^\prime}a_{r,c^\prime} } \cdot χ_{rel} \right] \end{equation}
\(χ_{min}\) = Minimum crop share allowed in the spatial unit
\(χ_{rel}\) = Minimum relative crop share – defines heterogeneity of crop shares for a crop in a spatial unit